Clustering evaluation
This time I did clustering with DBSCAN and HDBSCAN. These are fairly simple yet effective clustering algorithms suitable for any kind of data. Additionally, I’ve implemented a method evaluating the results of clustering with variety of metrics: Adjusted Mutual Information Score, Adjusted Random Index, Completeness, Homogeneity, Silhouette Coefficient, and V-measure. Finally, I did loads of experiments to pick te best parameters for those.
Choosing the best parameters settings
A snippet to produce results of clustering for variety of parameters settings.
import numpy as np
import pandas as pd
from cuckooml import Loader
from cuckooml import ML
loader = Loader()
loader.load_binaries("../../sample_data/dict")
ml = ML()
ml.load_simple_features(loader.get_simple_features())
ml.load_features(loader.get_features())
ml.load_labels(loader.get_labels())
filtered_features = ml.filter_dataset(ml.features)
results = {}
# full data
for eps in np.arange(0.1, 20.1, .1):
for ms in range(1, 21):
c = ml.cluster_dbscan(ml.features, eps, ms)
results["full_data+dbscan+eps="+str(eps)+"+ms="+str(ms)] = \
ml.assess_clustering(c, ml.labels, ml.features)
# filter noisy labels
c.columns=["cluster"]
clean = pd.concat([c, ml.labels], axis=1)
clean = clean[clean.cluster != -1]
cl = clean[["label"]]
cc = clean[["cluster"]]
cc.columns=["label"]
results["full_data+dbscan+noise+eps="+str(eps)+"+ms="+str(ms)] = \
ml.assess_clustering(cc, cl)
c = ml.cluster_dbscan(filtered_features, eps, ms)
results["filtered_data+dbscan+eps="+str(eps)+"+ms="+str(ms)] = \
ml.assess_clustering(c, ml.labels, filtered_features)
# filter noisy labels
c.columns=["cluster"]
clean = pd.concat([c, ml.labels], axis=1)
clean = clean[clean.cluster != -1]
cl = clean[["label"]]
cc = clean[["cluster"]]
cc.columns=["label"]
results["filtered_data+dbscan+noise+eps="+str(eps)+"+ms="+str(ms)] = \
ml.assess_clustering(cc, cl)
for ms in [None]+range(1, 21):
for mcs in range(2, 21):
c = ml.cluster_hdbscan(ml.features, ms, mcs)
results["full_data+hdbscan+ms="+str(ms)+"+mcs="+str(mcs)] = \
ml.assess_clustering(c, ml.labels, ml.features)
# filter noisy labels
c.columns=["cluster"]
clean = pd.concat([c, ml.labels], axis=1)
clean = clean[clean.cluster != -1]
cl = clean[["label"]]
cc = clean[["cluster"]]
cc.columns=["label"]
results["full_data+hdbscan+noise+ms="+str(ms)+"+mcs="+str(mcs)] = \
ml.assess_clustering(cc, cl)
c = ml.cluster_hdbscan(filtered_features, ms, mcs)
results["filtered_data+hdbscan+ms="+str(ms)+"+mcs="+str(mcs)] = \
ml.assess_clustering(c, ml.labels, filtered_features)
# filter noisy labels
c.columns=["cluster"]
clean = pd.concat([c, ml.labels], axis=1)
clean = clean[clean.cluster != -1]
cl = clean[["label"]]
cc = clean[["cluster"]]
cc.columns=["label"]
results["filtered_data+hdbscan+noise+ms="+str(ms)+"+mcs="+str(mcs)] = \
ml.assess_clustering(cc, cl)
results_df = pd.DataFrame(results).T
results_df.to_csv("clustering_results.csv")then the following code was used to produce figures and results placed below:
def vd(data, labels, clusters, learning_rate=200,
fig_name="custom"):
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from sklearn.manifold import TSNE
tsne = TSNE(learning_rate=learning_rate)
tsne_fit = tsne.fit_transform(data)
tsne_df = pd.DataFrame(tsne_fit, index=data.index, columns=['0', '1'])
tsne_dfl = pd.concat([tsne_df, labels], axis=1)
tsne_dfc = pd.concat([tsne_df, clusters], axis=1)
sns.lmplot("0", "1", data=tsne_dfl, fit_reg=False, hue="label",
scatter_kws={"marker":"D", "s":50}, legend_out=True)
plt.title(fig_name + " (lr:" + str(learning_rate) + ")")
plt.savefig(fig_name + "_gt_" + str(learning_rate) + ".png",
bbox_inches='tight', pad_inches=1.)
plt.close()
sns.lmplot("0", "1", data=tsne_dfc, fit_reg=False, hue="label",
scatter_kws={"marker":"D", "s":50}, legend_out=True)
plt.title(fig_name + " (lr:" + str(learning_rate) + ")")
plt.savefig(fig_name + "_cl_" + str(learning_rate) + ".png",
bbox_inches='tight', pad_inches=1.)
plt.close()
new_res = {}
for i in results:
if "noise" not in i:
new_res[i] = results[i]
nr = pd.DataFrame(new_res).T
# Then e.g.
nr[nr["Homogeneity"] > 0.1][["Homogeneity"]]
sorted(set(nr["Homogeneity"].tolist()))[-100:]
# And to plot
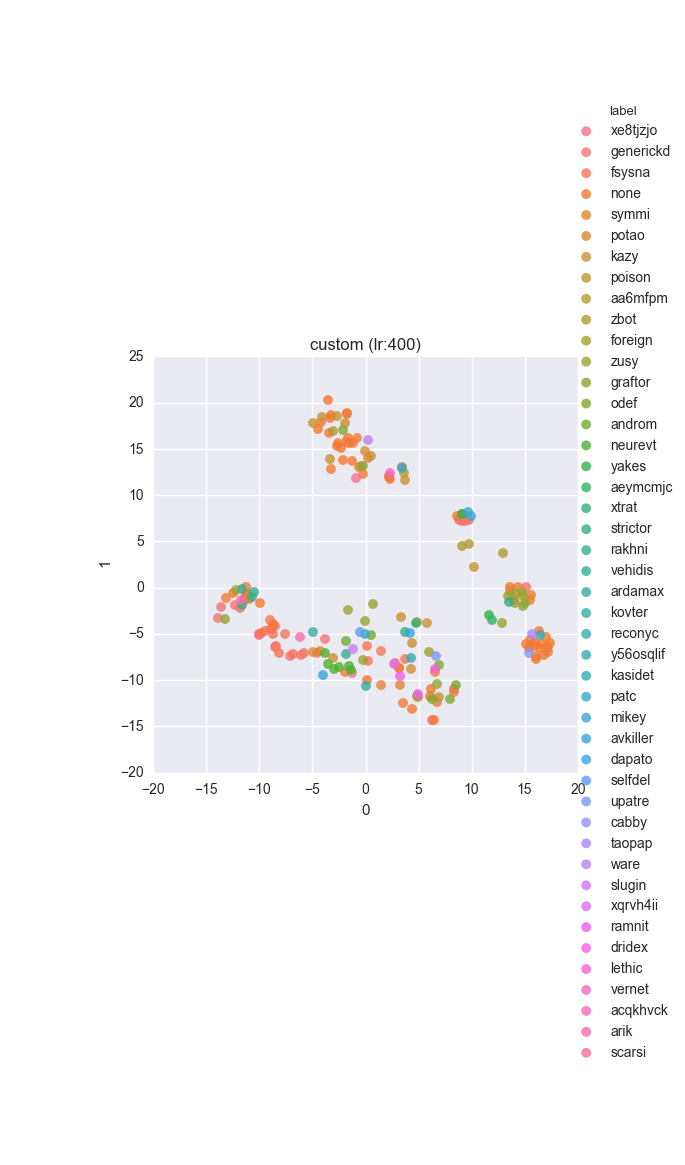
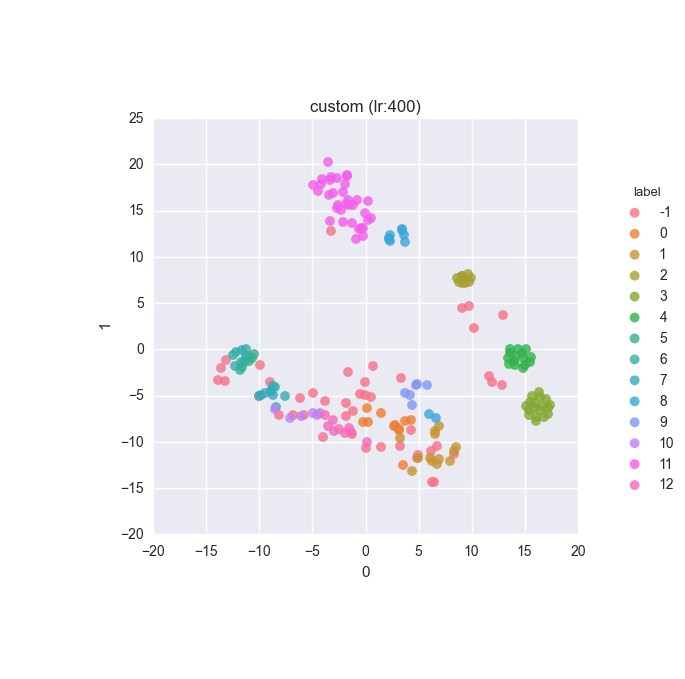
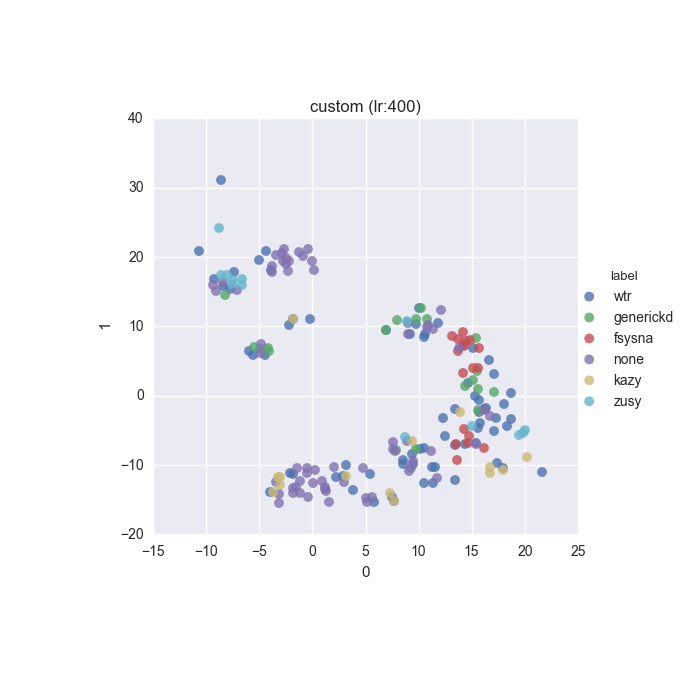
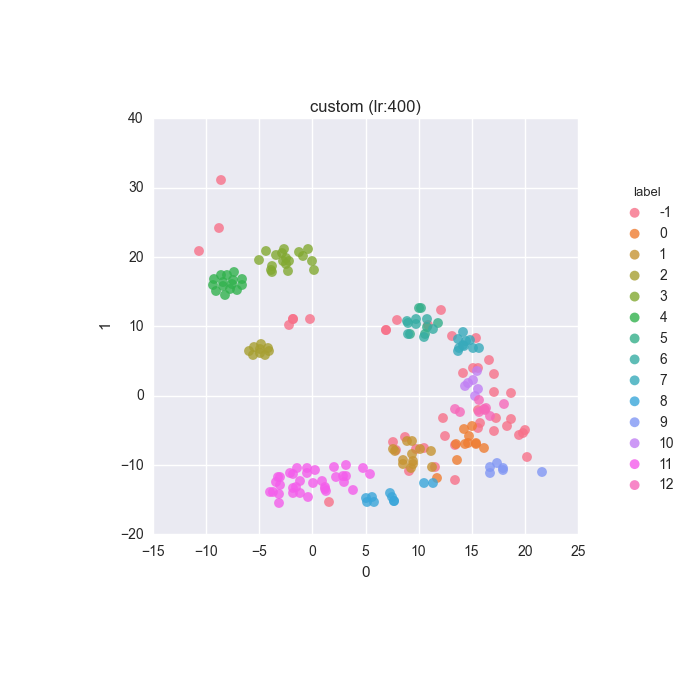
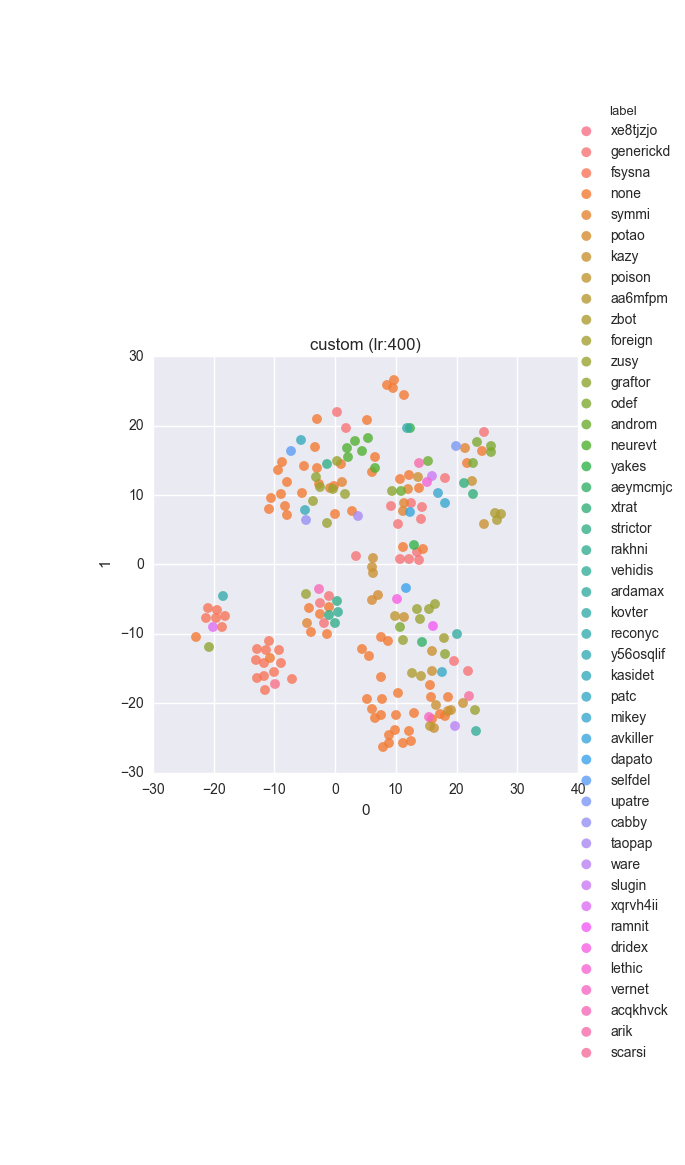
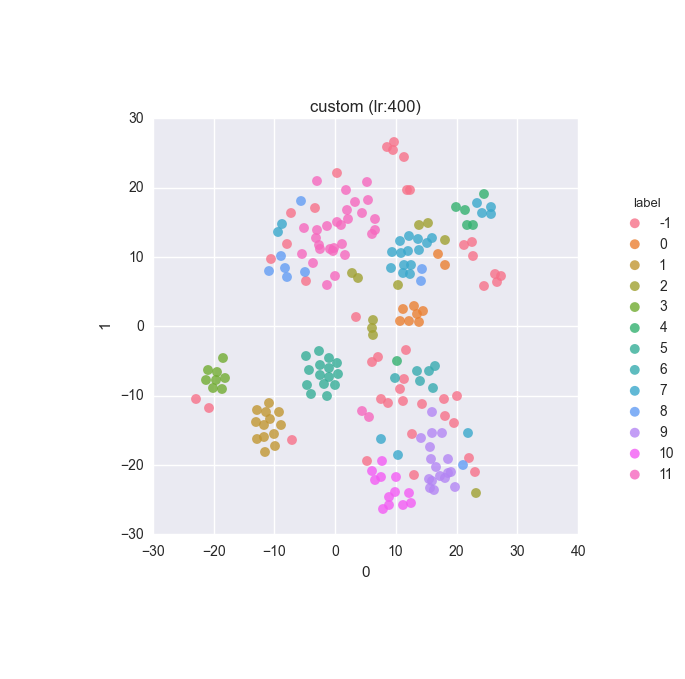
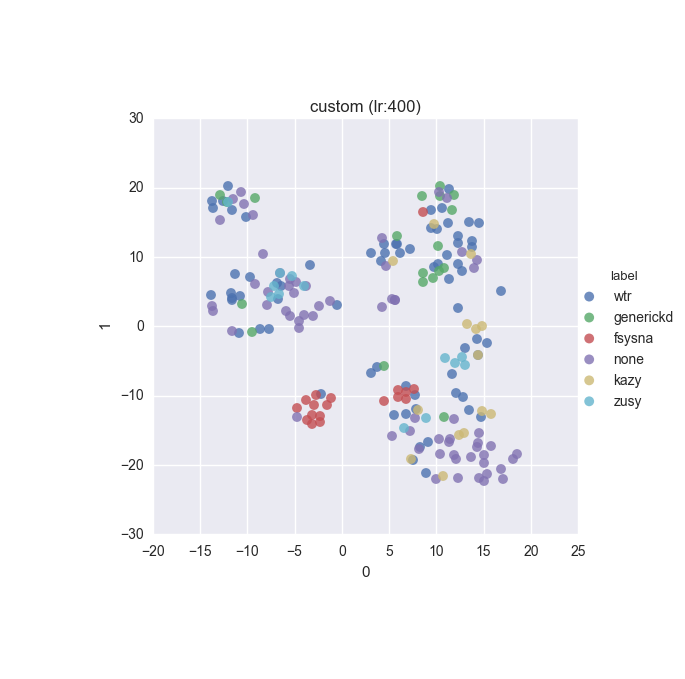
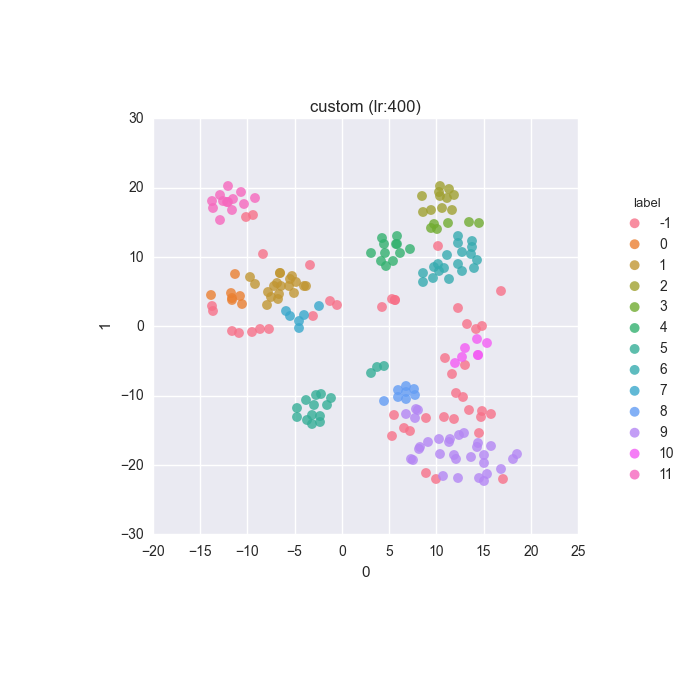
vd(filtered_features, ml.labels, ml.cluster_hdbscan(filtered_features, 1, 6), learning_rate=400)Results
After all kind of analyses with min_samples=1.0 and min_cluster_size=6 HDBSCAN seems to do quite well on the dataset with full set of features. It does not outperform the rest in all of the cases but provides top-of-the-ranking performance with reasonable cluster number and cluster structure. Below is table with results for various dataset combination:
- full - the full set of features
- filtered - only the features that are denser than 10%
- extended - the original set of features
- simple - the simple set of features
- relabelled - labels with less than 10 instances removed from the ground truth
Clustering performance metrics
| Data type | Adjusted Mutual Information | Adjusted Random Index | Completeness | Homogeneity | Silhouette Coefficient | V-measure |
|---|---|---|---|---|---|---|
| filtered_extended | 0.217 | 0.107 | 0.497 | 0.436 | 0.305 | 0.465 |
| filtered_extended_relabelled | 0.142 | 0.079 | 0.208 | 0.326 | 0.305 | 0.254 |
| filtered_simple | 0.240 | 0.136 | 0.518 | 0.445 | 0.331 | 0.478 |
| filtered_simple_relabelled | 0.161 | 0.078 | 0.222 | 0.340 | 0.331 | 0.268 |
| full_extended | 0.217 | 0.107 | 0.497 | 0.436 | 0.305 | 0.465 |
| full_extended_relabelled | 0.142 | 0.079 | 0.208 | 0.326 | 0.305 | 0.254 |
| full_simple | 0.240 | 0.136 | 0.518 | 0.445 | 0.331 | 0.478 |
| full_simple_relabelled | 0.161 | 0.078 | 0.222 | 0.340 | 0.331 | 0.268 |
Clustering visualisations
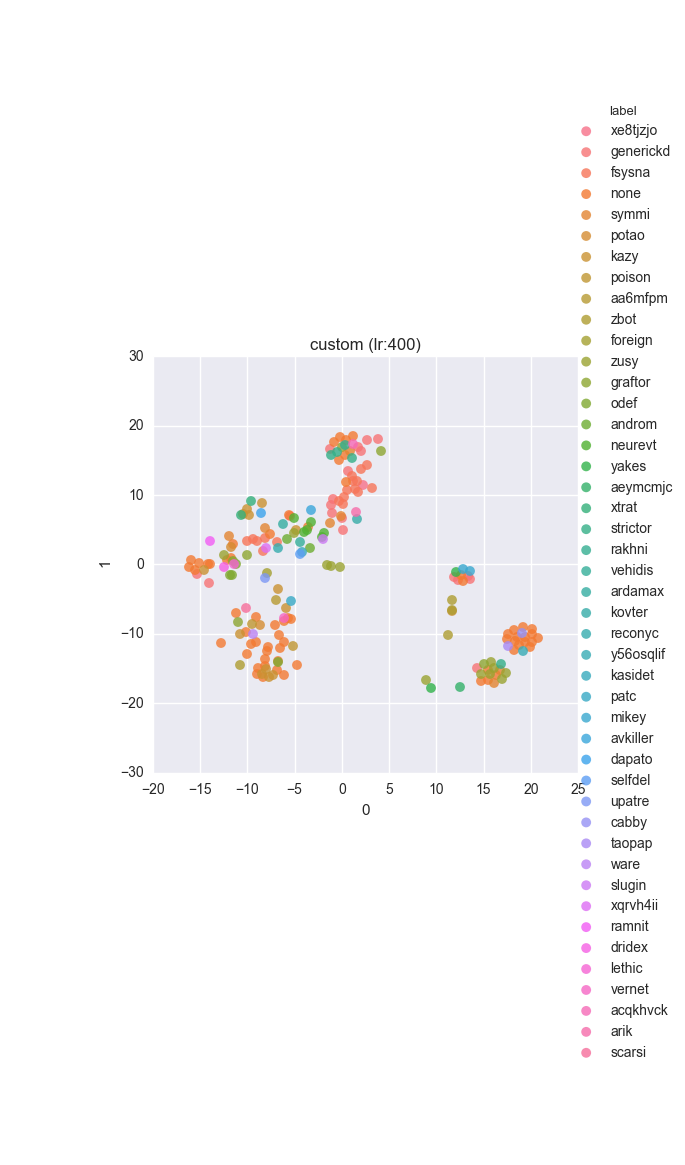
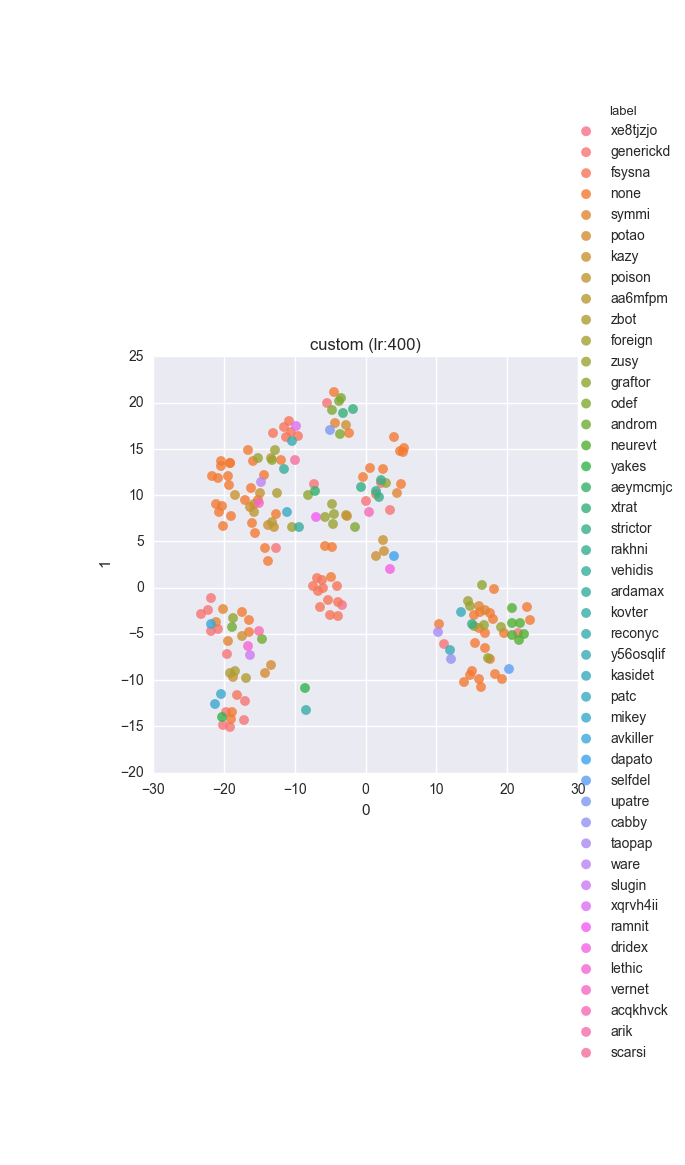
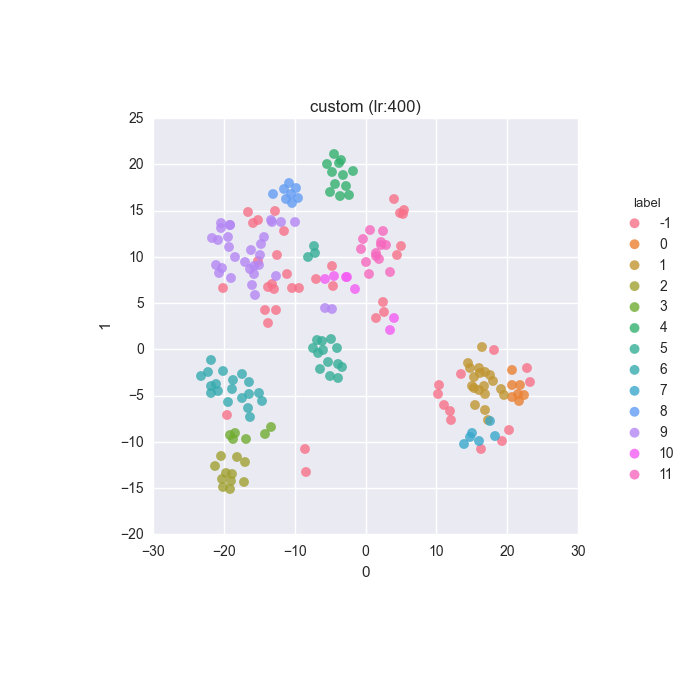
Extended dataset - full feature set - ground truth
Extended dataset - full feature set - clustering results
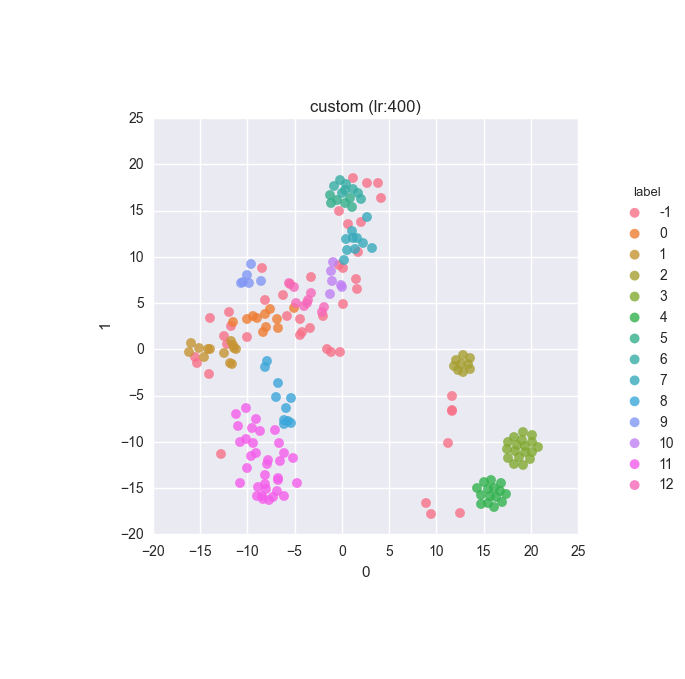
Extended dataset - full feature set - relabelled - ground truth
Extended dataset - full feature set - relabelled - clustering results
Extended dataset - filtered full feature set - ground truth
Extended dataset - filtered full feature set - clustering results
Extended dataset - filtered full feature set - relabelled - ground truth
Extended dataset - filtered full feature set - relabelled - clustering results
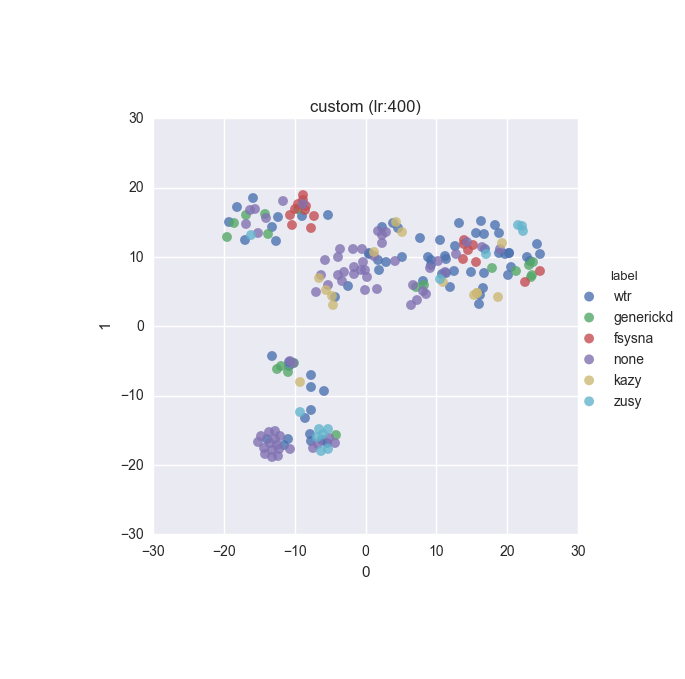
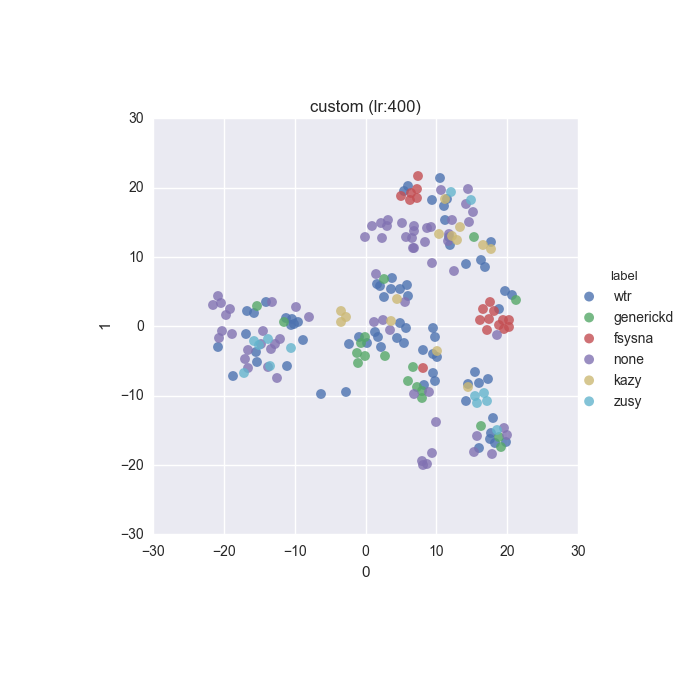
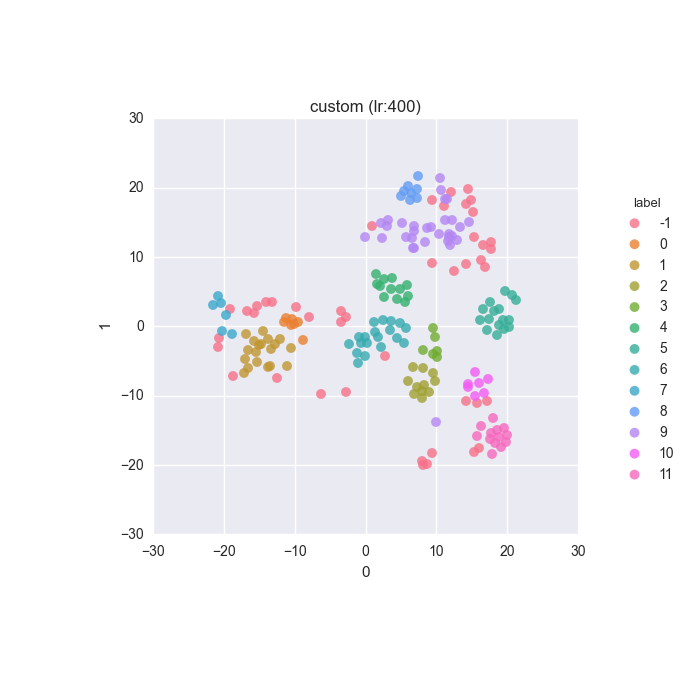
Simple dataset - full feature set - ground truth
Simple dataset - full feature set - clustering results
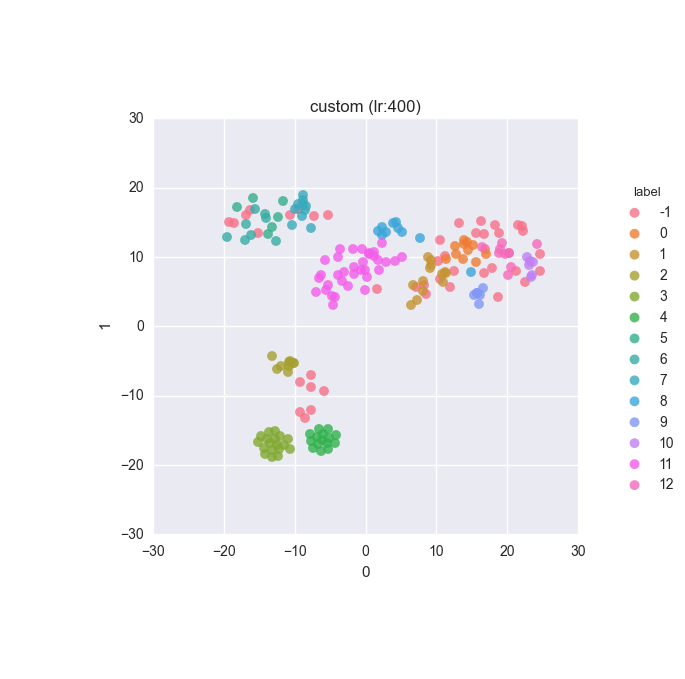
Simple dataset - full feature set - relabelled - ground truth
Simple dataset - full feature set - relabelled - clustering results
Simple dataset - filtered feature set - ground truth
Simple dataset - filtered feature set - clustering results
Simple dataset - filtered feature set - relabelled - ground truth
Simple dataset - filtered feature set - relabelled - clustering results
Future work
For the near future I’d like to test this approach for more data and possibly try out some other clustering algorithms.
It seems like the next natural step is novelity and anomaly detection. It might be worth investigating whether it’s possible to somehow get the probabilities out of HDBSCAN to facilitate this.