About malware features
For the past two weeks I’ve been experimenting with various malware feature transformations and combinations. It came to my understanding that using any Euclidean distance-based clustering for a dataset where more than 95% of boolean features might not be the best approach.
In our case there is significantly more features than data samples what usually has a detrimental effect on clustering. # data points » # features is usually advisable, with the number of features being around 10% of the number of samples for the best effects. If this does not hold noisy features can be overwhelming for clustering especially when some clusters are underrepresented in the data. Some of the clustering possibilities are presented below.
Full feature
Using the full feature set (19187 features) gives us 57 outliers (out of 199 malware samples) with clusters fit being (HDBSCAN: min_cluster_size=5, min_samples=1):
| Adjusted Random Index | 0.203094 |
| Adjusted Mutual Information Score | 0.338447 |
| Completeness | 0.563312 |
| Silhouette Coefficient | -0.197215 |
| V-measure | 0.556734 |
| Homogeneity | 0.550308 |
and clusters and outliers fit:
| Adjusted Random Index | 0.082914 |
| Adjusted Mutual Information Score | 0.212254 |
| Completeness | 0.504984 |
| Silhouette Coefficient | 0.216329 |
| V-measure | 0.465022 |
| Homogeneity | 0.430921 |
and not so good clusters when inspected closer.
Simple feature
Simple features have 61 features and 36 outliers (HDBSCAN: min_cluster_size=5, min_samples=1) with clustering fit:
| Adjusted Random Index | 0.133554 |
| Adjusted Mutual Information Score | 0.269060 |
| Completeness | 0.522440 |
| Silhouette Coefficient | -0.124457 |
| V-measure | 0.501318 |
| Homogeneity | 0.481838 |
and clustering with outliers fit:
| Adjusted Random Index | 0.089312 |
| Adjusted Mutual Information Score | 0.214820 |
| Completeness | 0.489151 |
| Silhouette Coefficient | 0.269974 |
| V-measure | 0.462284 |
| Homogeneity | 0.438215 |
and again not so good clusters when inspected closer.
Numerical features
There are 21 numerical features 21, with clustering (HDBSCAN: min_cluster_size=7, min_samples=1) resulting in 49 outliers and cluster fit:
| Adjusted Random Index | 0.164326 |
| Adjusted Mutual Information Score | 0.263059 |
| Completeness | 0.502374 |
| Silhouette Coefficient | -0.607331 |
| V-measure | 0.479650 |
| Homogeneity | 0.458894 |
and clustering + oultiers fit:
| Adjusted Random Index | 0.108847 |
| Adjusted Mutual Information Score | 0.199091 |
| Completeness | 0.490092 |
| Silhouette Coefficient | 0.276731 |
| V-measure | 0.437520 |
| Homogeneity | 0.395134 |
and again not so good clusters when inspected closer. Nevertheless, very good results when considering the results and the number of features used.
Simple features: reduced dimensionality
I have also experimented with variety of combination of these: using log binning for the numerical features and various thresholding to delete the sparse feature; some of them resulted in quite good clustering with the best result achieved by simple features where only features covering more than 10% of the examples were used (17 features). Clustering (HDBSCAN: min_cluster_size=5, min_samples=1) resulted in 57 outliers with the cluster fit statistics:
| Adjusted Random Index | 0.230443 |
| Adjusted Mutual Information Score | 0.363688 |
| Completeness | 0.588787 |
| Silhouette Coefficient | -0.175176 |
| V-measure | 0.575281 |
| Homogeneity | 0.562382 |
and clustering + outliers statistics:
| Adjusted Random Index | 0.107736 |
| Adjusted Mutual Information Score | 0.230274 |
| Completeness | 0.516208 |
| Silhouette Coefficient | 0.326374 |
| V-measure | 0.478756 |
| Homogeneity | 0.446371 |
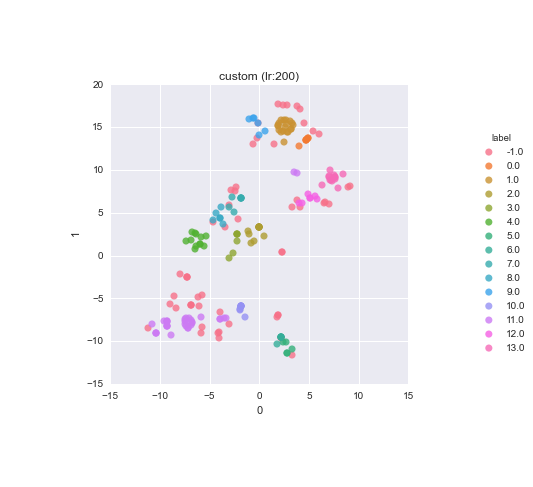
Clusters visualisation
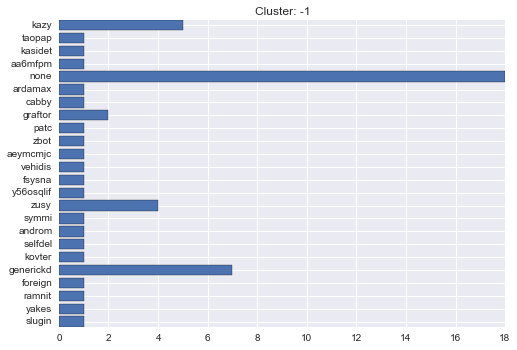
-1 cluster statistics
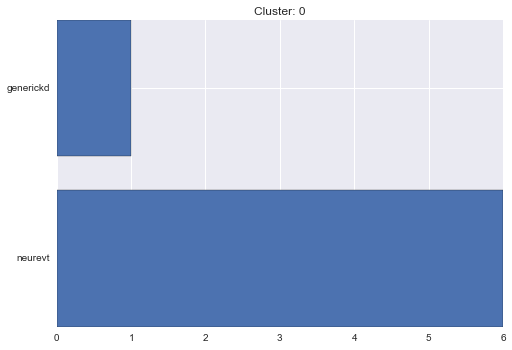
0 cluster statistics
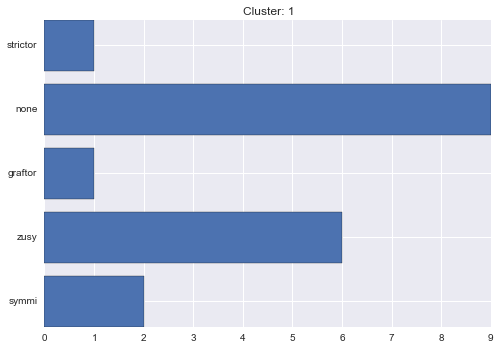
1 cluster statistics
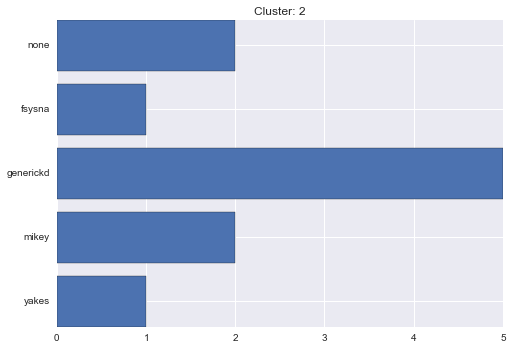
2 cluster statistics
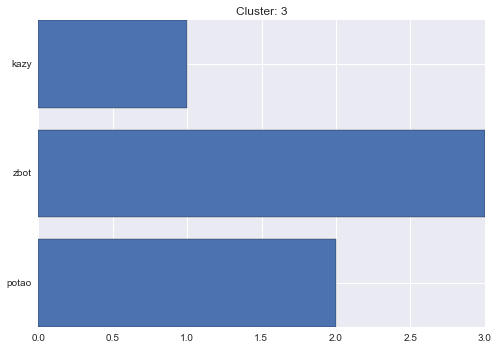
3 cluster statistics
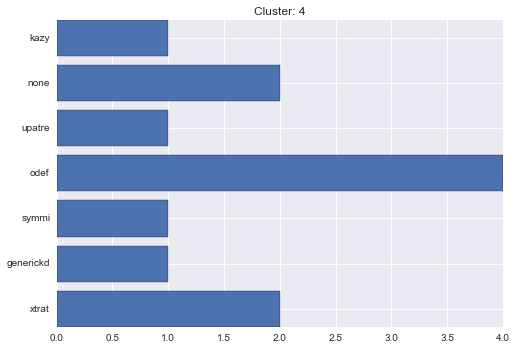
4 cluster statistics
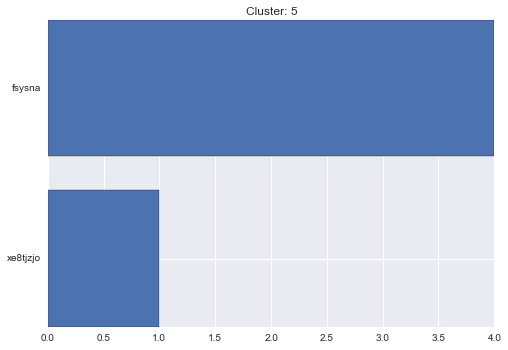
5 cluster statistics
6 cluster statistics
7 cluster statistics
8 cluster statistics
9 cluster statistics
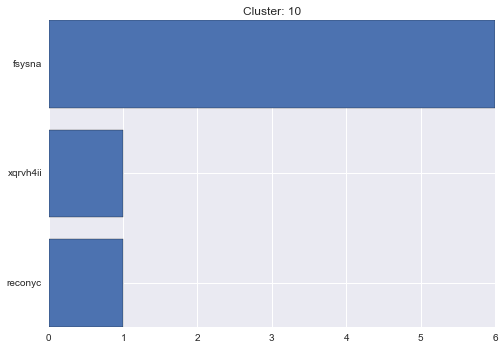
10 cluster statistics
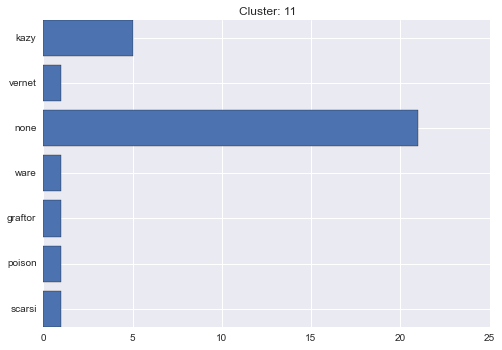
11 cluster statistics
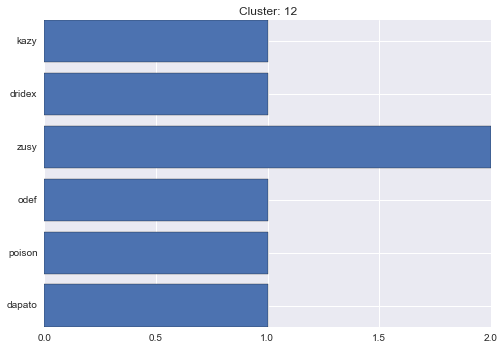
12 cluster statistics
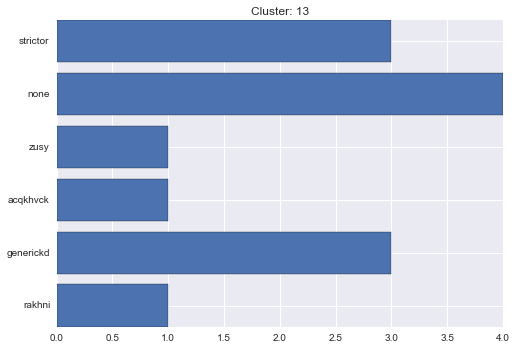
13 cluster statistics
Summary
To sum up, we have good features but they are underrepresented in the simple dataset used for the development of cuckoml; to many samples are healthy (none label). Furthermore, the problem with combining binary (vast majority) with all the rest of the features causes them to become far apart, hence, undermines the clustering mechanisms; unfortunately, applying log-binning to the numerical features is of little help here.