Clustering misc
I spent the last couple of days to implement miscellaneous functions helping with clustering. Among others: saving clustering results into JSON, label statistics per cluster, comparing a new sample to in-memory clustering and basic version of anomaly detection.
Moreover, I’ve started developing a Jupyter Notebook to showcase all the implemented code during GSoC’16.
Clustering statistics
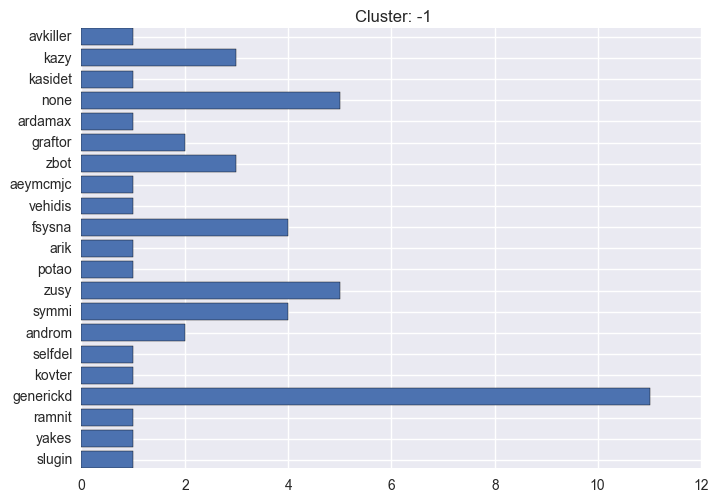
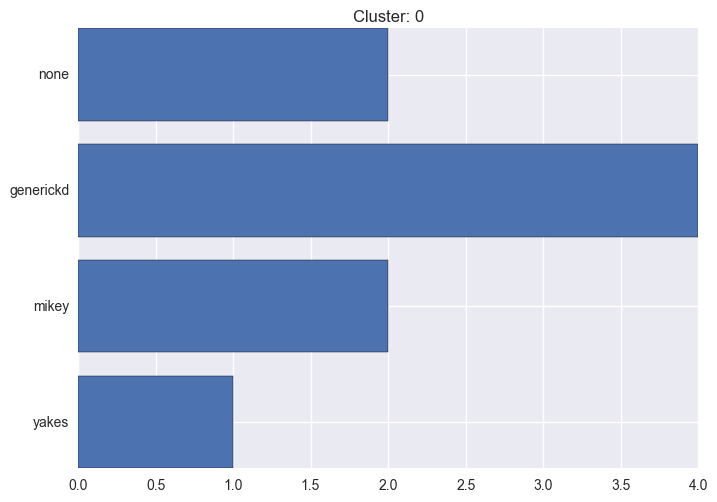
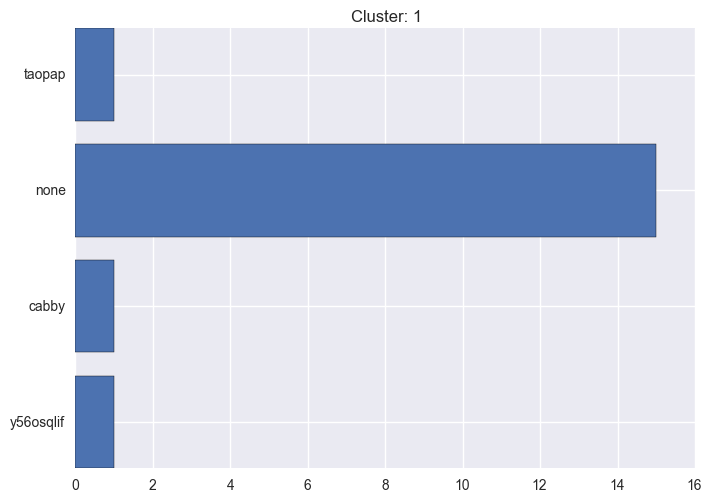
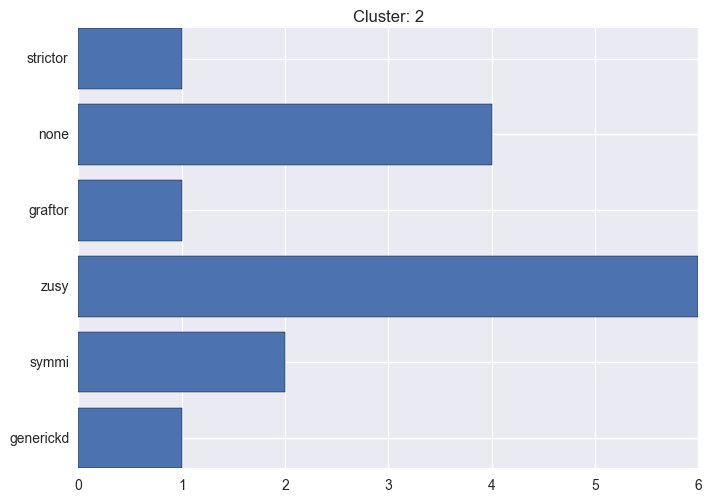
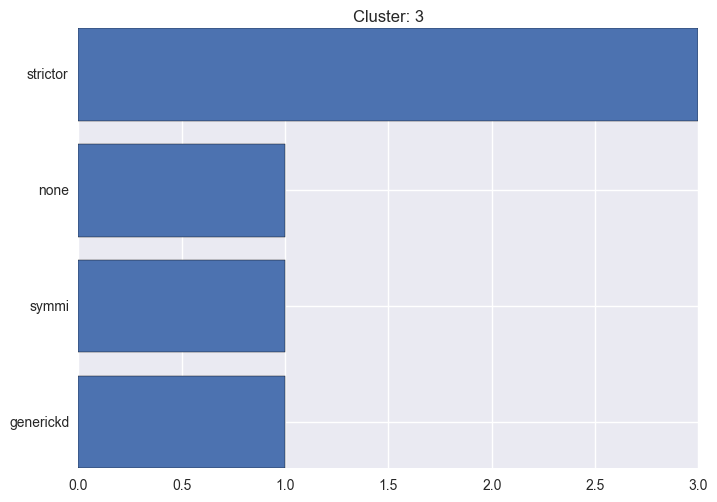
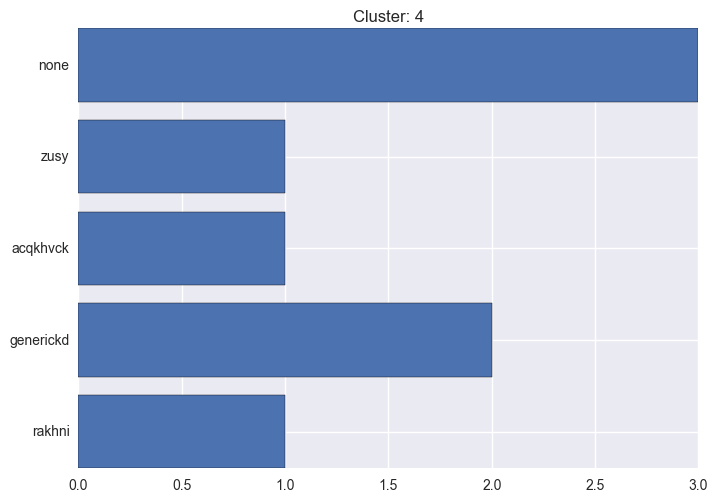
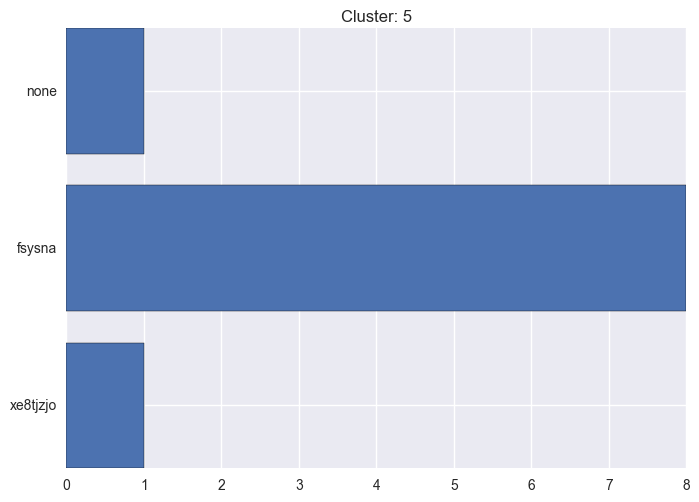
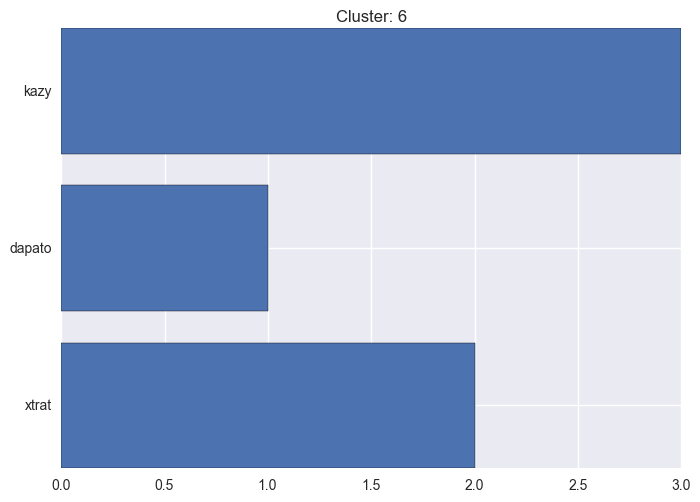
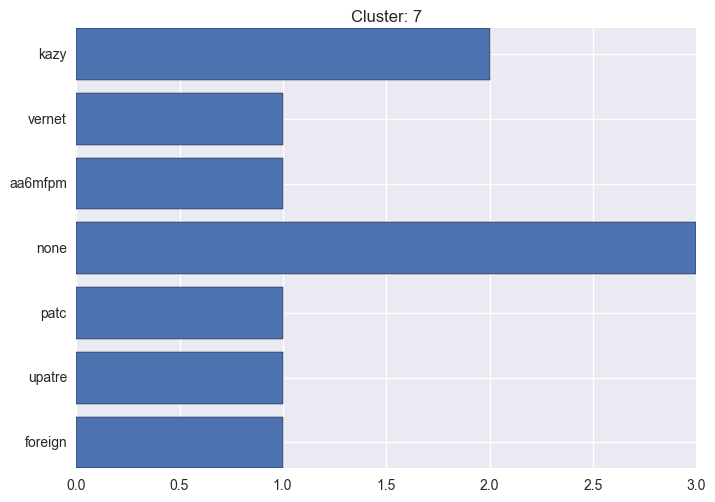
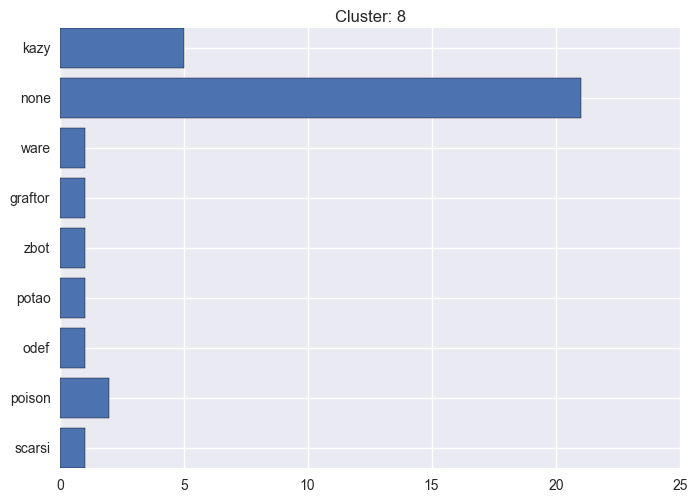
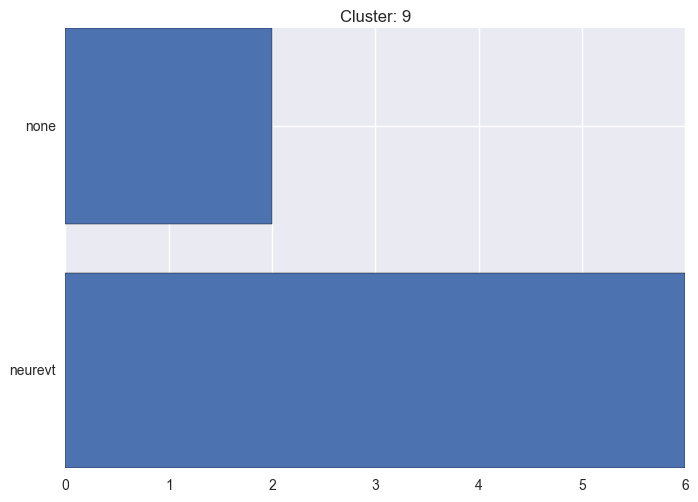
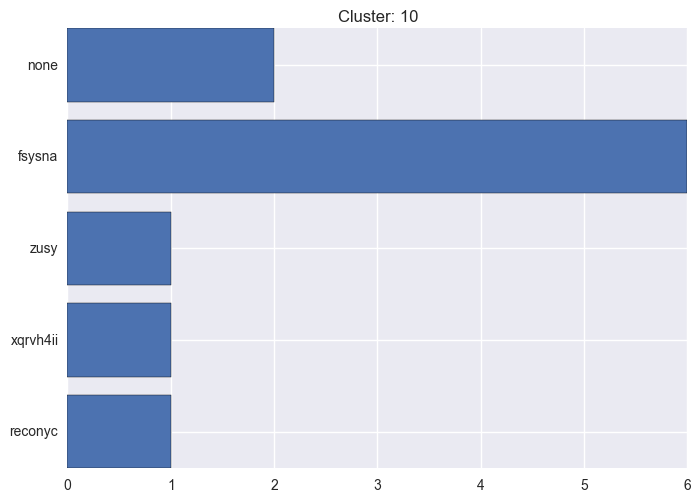
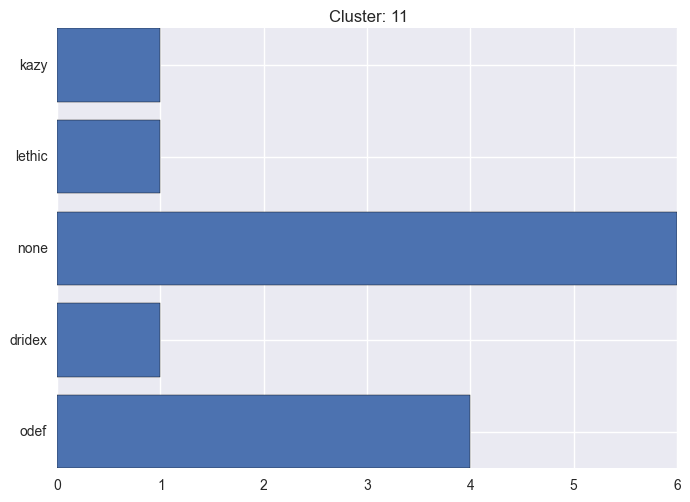
It seems worth discovering per cluster structure. The easiest way is to produce a bar plot that counts ground truth (VirusTotal) labels per discovered cluster.
To get this kind of plots
A snipped to produce results of clustering for variety of parameters settings.
from cuckooml import Loader
from cuckooml import ML
loader = Loader()
loader.load_binaries("../../sample_data/dict")
ml = ML()
ml.load_simple_features(loader.get_simple_features())
ml.load_features(loader.get_features())
ml.load_labels(loader.get_labels())
ml.cluster_hdbscan(ml.features, 1, 6)
# assess clustering fit (various metrics - see previous post)
ml.assess_clustering(ml.clustering["hdbscan"]["clustering"], ml.labels, ml.features)
# get label statistics per cluster
ml.clustering_label_distribution(ml.clustering["hdbscan"]["clustering"], ml.labels, True)See the plots below for the visualisations.
-1 cluster statistics
0 cluster statistics
1 cluster statistics
2 cluster statistics
3 cluster statistics
4 cluster statistics
5 cluster statistics
6 cluster statistics
7 cluster statistics
8 cluster statistics
9 cluster statistics
10 cluster statistics
11 cluster statistics
Updating JSONs
To update the JSONs with clustering results use the following code:
from cuckooml import Loader
from cuckooml import ML
loader = Loader()
loader.load_binaries("../../sample_data/dict")
ml = ML()
ml.load_simple_features(loader.get_simple_features())
ml.load_features(loader.get_features())
ml.load_labels(loader.get_labels())
# perform clustering and save the results in-memory
ml.cluster_hdbscan(ml.features, 1, 6)
ml.save_clustering_results(loader, "../../sample_data/dict_cluster")Compare a new sample to already fitted data
To compare a new sample to alredy existing clustering you can use the code snipped given below. It will return cluster ID, cluster membership probability, and outlier score.
from cuckooml import Instance
from cuckooml import Loader
from cuckooml import ML
loader = Loader()
loader.load_binaries("../../sample_data/dict")
ml = ML()
ml.load_simple_features(loader.get_simple_features())
ml.load_features(loader.get_features())
ml.load_labels(loader.get_labels())
ml.cluster_hdbscan(ml.features)
# get a new sample
new_sample = Instance()
# get a new sample and save it in-memory as *5_new*
new_sample.load_json("../../sample_data/dict/5", "5_new")
new_sample.label_sample()
new_sample.extract_features()
new_sample.extract_basic_features()
# compare the new sample
ml.compare_sample(new_sample)Anomaly detection
Anomaly detection for new malware - especially in clustering scenario - is a complicated task. The first attempt (and implementation) returns anomalies considered in variety of aspects.
At the moment these are: outliers detected by HDBSCAN algorithm, samples with high outlier score, elements from clusters that are not homogeneous, and per cluster samples with low probability of belonging to that cluster.
from cuckooml import Loader
from cuckooml import ML
loader = Loader()
loader.load_binaries("../../sample_data/dict")
ml = ML()
ml.load_simple_features(loader.get_simple_features())
ml.load_features(loader.get_features())
ml.load_labels(loader.get_labels())
ml.cluster_hdbscan(ml.features)
ml.anomaly_detection()cuckooml showcase (Jupyter Notebook)
Finally, I decided to pull together all of the code snippets placed on the blog so far into one Jupyter Notebook. I think that this will be a great way to showcase cuckooml capabilities. Additionally, it is incredibly easy way to introduce new users and potential contributors to the project. I’ll publish a new post dedicated to this topic anytime soon.